Webinar: Presentation of GeVarLi, a public health tool developed for AFROSCREEN project
Webinar: Presentation of GeVarLi, a public health tool developed for AFROSCREEN project
The third webinar organized under the AFROSCREEN project was held by Nicolas Fernandez, bioinformatics engineer from UMI 233 TransVIHMI IRD (Institut de recherche pour le développement – Montpellier). He presented the principles of reproducibility, F.A.I.R, Open Science and traceability of bioinformatics analyses, applied to the genomic analysis of SARS-CoV-2 variants, through the GeVarLi workflow (a Snakemake pipeline for genome assembly), variant calling and lineage assignment for SARS-CoV-2 and other emerging viruses.

Nicolas Fernandez talked about the crisis of reproducibility affecting several scientific fields but also about several principles and good practices allowing to answer this observation. All research should follow the F.A.I.R. principle, presenting data asFindable,Accessible,Interoperable andReusable, as well as the notion ofOpen Science.
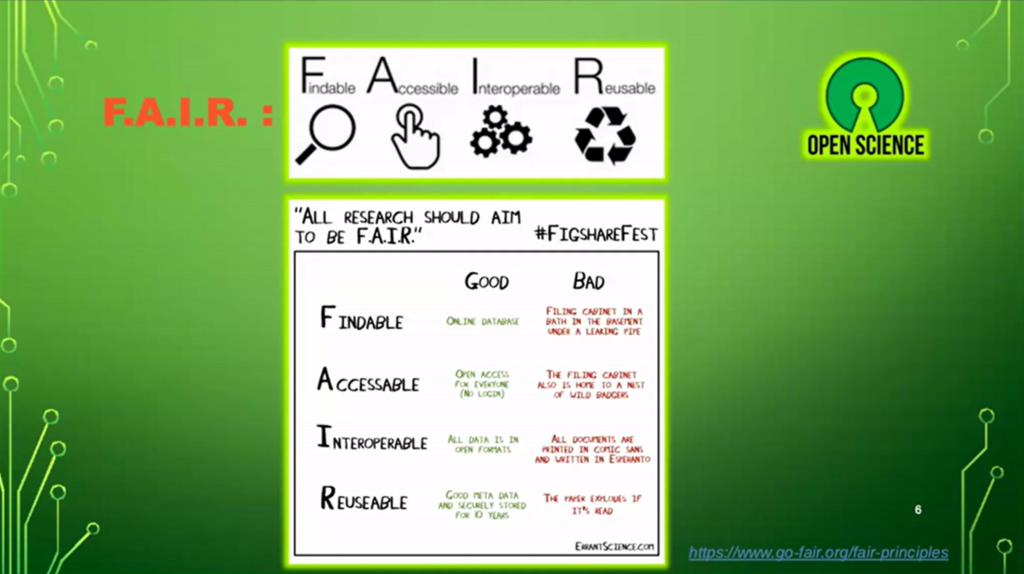
Nicolas Fernandez nous a rappelé l’importance d’appliquer ces principes à nos analyses bio-informatiques, avec l’utilisation de divers outils, comme les gestionnaires de F.A.I.R. workflows (such as Snakemake, Nextflow), theenvironments (such as Anaconda, Docker) and the need to publish and track (versioning) one’s code with tools such as Git (including IRD’s GitLab instance, IRDForge).
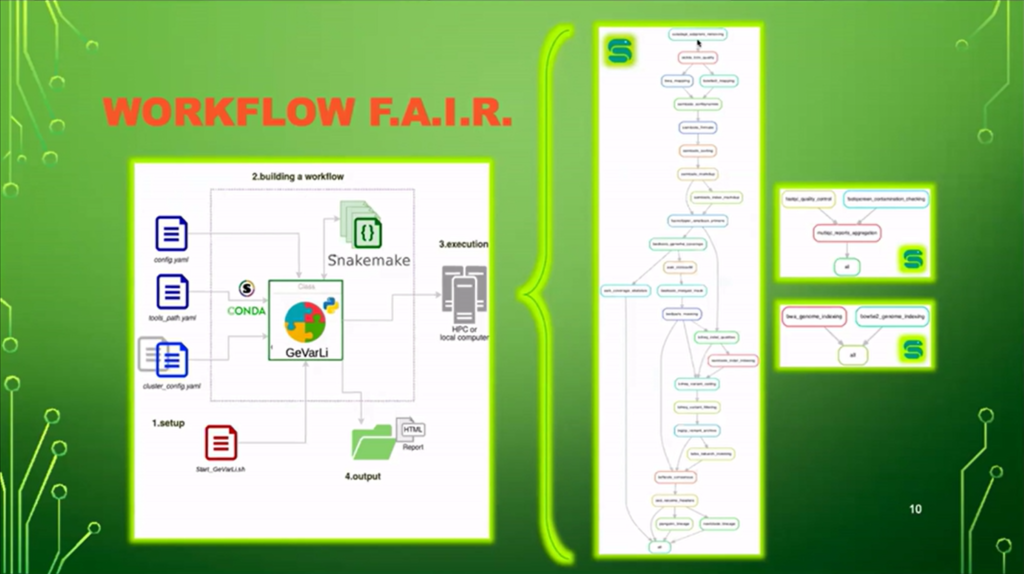
Nicolas Fernandez then presented an example of F.A.I.R. workflow with the ” GeVarLi : GEnomeassembly, VARiantcalling and LIneageassignment” tool, an automated pipeline in Snakemake developed for the genomic analysis of SARS-CoV-2 and other emerging viruses He presented us its genesis, where to find it, how to install and use it as well as the results obtained and its advantages.

Nicolas Fernandez insisted in particular on the fact that the workflow was open, free, scalable, and open to contribution and collaborative participation, as well as the importance of citing and thanking the authors of free tools created by and for the community.
To learn more, watch the entire webinar:
- FR : https://youtu.be/haBVJdWqNeE
- EN : https://youtu.be/ssEzpN2NkZc
- and more information on GeVarli: https://forge.ird.fr/transvihmi/nfernandez/GeVarLi
Nicolas Fernandez talked about the crisis of reproducibility affecting several scientific fields but also about several principles and good practices allowing to answer this observation. All research should follow the F.A.I.R. principle, presenting data asFindable,Accessible,Interoperable andReusable, as well as the notion ofOpen Science.

Nicolas Fernandez nous a rappelé l’importance d’appliquer ces principes à nos analyses bio-informatiques, avec l’utilisation de divers outils, comme les gestionnaires de F.A.I.R. workflows (such as Snakemake, Nextflow), theenvironments (such as Anaconda, Docker) and the need to publish and track (versioning) one’s code with tools such as Git (including IRD’s GitLab instance, IRDForge).

Nicolas Fernandez then presented an example of F.A.I.R. workflow with the ” GeVarLi : GEnomeassembly, VARiantcalling and LIneageassignment” tool, an automated pipeline in Snakemake developed for the genomic analysis of SARS-CoV-2 and other emerging viruses He presented us its genesis, where to find it, how to install and use it as well as the results obtained and its advantages.

Nicolas Fernandez insisted in particular on the fact that the workflow was open, free, scalable, and open to contribution and collaborative participation, as well as the importance of citing and thanking the authors of free tools created by and for the community.
To learn more, watch the entire webinar:
- FR : https://youtu.be/haBVJdWqNeE
- EN : https://youtu.be/ssEzpN2NkZc
- and more information on GeVarli: https://forge.ird.fr/transvihmi/nfernandez/GeVarLi